The SeaClass R Package
The Operations Technology and Advanced Analytics Group (OTAAG) at Seagate Technology has decided to share an internal project that helps accelerate development of classification models. The interactive SeaClass tool is contained in an R-based package built using shiny and other CRAN packages commonly used for binary classification. The package is free to use and develop further, but any analysis mistakes are the sole responsibility of the user. Check out the demo video below:
Origin Story
Manufacturer data sets used for pass/fail analysis are typically highly imbalanced with many more passing cases than failing cases. In some situations, the failure examples may be scarce or nonexistent. In these extreme cases complex modeling techniques are likely inadvisable. Data scientists commonly consider data scarcity, class imbalance, and data dimensionality when discriminating between competing candidate approaches, such as anomaly detection, simple models, and complex models. Standard approaches are easily identified within each of these analysis categories, and can be exploited as reasonable initial classification analysis baselines.
The SeaClass application was created to generate template R code for the commonly encountered classification problems described above. The application allows data analysts to explore multiple models quickly with essentially no programming required. SeaClass provides an option to download the corresponding R code for further model training/testing. This workflow enables our data analysts to jump-start their modeling, saving time and initial hassles.
The Advanced Analytics group decided to open-source the package for several reasons. Firstly, we encourage other contributors to suggest improvements to the SeaClass application. Additionally, we are hopeful our application will inspire other code-generating projects within the R community. Lastly, our group benefits greatly from open-source tools, and it’s nice to give back every once in a while.
Package Overview
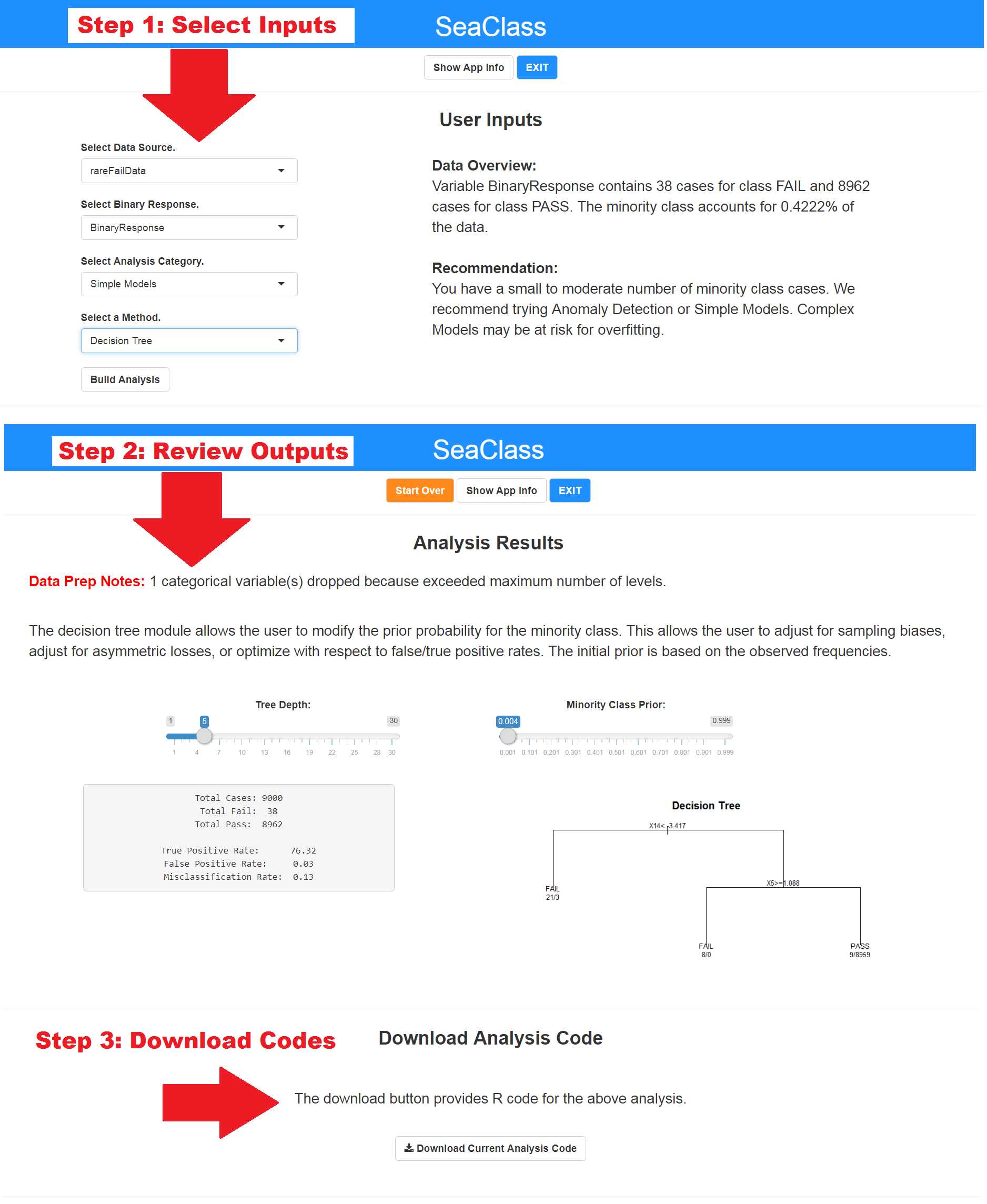
The SeaClass R package provides tools for analyzing classification problems. In particular, specialized tools are available for addressing the problem of imbalanced data sets. The SeaClass application provides an easy-to-use interface that requires only minimal R programming knowledge to get started, and can be launched using the RStudio Addins menu. The application allows the user to explore numerous methods by simply clicking on the available options and interacting with the generated results. The user can choose to download the codes for any procedures they wish to explore further. SeaClass was designed to jump-start the analysis process for both novice and advanced R users. See screenshots below for one demonstration.
Installation Instructions
The SeaClass application depends on multiple R packages. To install SeaClass and its dependencies, run:
install.packages('devtools')
devtools::install_github('ChrisDienes/SeaClass')Usage Instructions
Step 1. Begin by loading and preparing your data in R. Some general advice:
- Your data set must be saved as an R data frame object.
- The data set must contain a binary response variable (0/1, PASS/FAIL, A/B, etc.)
- All other variables must be predictor variables.
- Predictor variables can be numeric, categorical, or factors.
- Including too many predictors may slow down the application and weaken performance.
- Categorical predictors are often ignored when the number of levels exceeds 10, since they tend to have improper influences.
- Missing values are not allowed and will throw a flag. Please remove or impute NAs prior to starting the app.
- Keep the number of observations (rows) to a medium or small size.
- Data sets with many rows (>10,000) or many columns (>30) may slow down the app’s interactive responses.
Step 2. After data preparation, start the application by either loading SeaClass from the RStudio Addins drop-down menu, or by loading the SeaClass function from the command line. For example:
library(SeaClass)
### Make some fake data:
X <- matrix(rnorm(10000,0,1),ncol=10,nrow=1000)
X[1:100,1:2] <- X[1:100,1:2] + 3
Y <- c(rep(1,100), rep(0,900))
Fake_Data <- data.frame(Y = Y , X)
### Load the SeaClass rare failure data:
data("rareFailData")
### Start the interactive GUI:
SeaClass()If the application fails to load, you may need to specify your favorite browser path first. For example:
options(browser = "C:/Program Files (x86)/Google/Chrome/Application/chrome.exe")Step 3. The user has various options for configuring their analysis within the GUI. Once the analysis runs, the user can view the results, interact with the results (module-dependent), save the underlying R script, or start over. Additional help is provided within the application. See above screenshots for one depiction of these steps.
Step 4. Besides the SeaClass function, several other functions are contained within the library. For example:
### List available functions:
ls("package:SeaClass")
### Note this is a sample data set:
# data(rareFailData)
### Note code_output is a support function for SeaClass, not for general use.
### View help:
?accuracy_threshold
### Run example from help file:
### General Use: ###
set.seed(123)
x <- c(rnorm(100,0,1),rnorm(100,2,1))
group <- c(rep(0,100),rep(2,100))
accuracy_threshold(x=x, group=group, pos_class=2)
accuracy_threshold(x=x, group=group, pos_class=0)
### Bagged Example ###
set.seed(123)
replicate_function = function(index){accuracy_threshold(x=x[index], group=group[index], pos_class=2)[[2]]}
sample_cuts <- replicate(100, {
sample_index = sample.int(n=length(x),replace=TRUE)
replicate_function(index=sample_index)
})
bagged_scores <- sapply(x, function(x) mean(x > sample_cuts))
unbagged_cut <- accuracy_threshold(x=x, group=group, pos_class=2)[[2]]
unbagged_scores <- ifelse(x > unbagged_cut, 1, 0)
# Compare AUC:
PRROC::roc.curve(scores.class0 = bagged_scores,weights.class0 = ifelse(group==2,1,0))[[2]]
PRROC::roc.curve(scores.class0 = unbagged_scores,weights.class0 = ifelse(group==2,1,0))[[2]]
bagged_prediction <- ifelse(bagged_scores > 0.50, 2, 0)
unbagged_prediction <- ifelse(x > unbagged_cut, 2, 0)
# Compare Confusion Matrix:
table(bagged_prediction, group)
table(unbagged_prediction, group)
You may leave a comment below or discuss the post in the forum community.rstudio.com.